My Cart
Your Shopping Cart is currently empty. Use Quick Order or Search to quickly add items to your order!
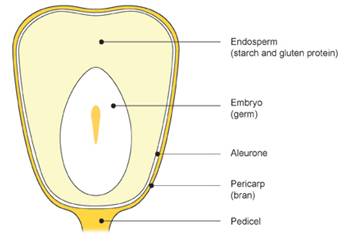 |
Figure 1 Parts of a corn kernel. |
David Micklos
DNA Learning Center, Cold Spring Harbor Laboratory
In most industrialized countries, people are used to eating fresh, canned, frozen, or dried products made from yellow or white corn. Corn, or maize as it is known to the rest of the world, is a wonderful model for genetic studies. An ear of maize has several hundred kernels—each of which is an embryo that arose from a unique fertilization event between a sperm and egg. Controlled genetic crosses are made by transfering pollen from one male plant tassel to the silks of an immature female cob. Each silk is a long style (elongated portion of the pistil) that conducts a sperm to the ovary inside each kernel (see Figure 1).
Each ear of corn holds the offspring of a single set of parents, providing a large family of siblings in which to study how traits pass from one generation to the next. (Imagine one couple having hundreds of children!) In many cases, each kernel needs to be fully grown for you to observe various traits, or phenotypes, in an adult plant. However, you can follow some traits simply by observing the kernels themselves.
 |
Figure 2 Ear of corn with pigmented and unpigmented kernels. |
Indian corn, used as decoration at Halloween and Thanksgiving, has a mixture of brightly colored kernels. The red, blue, and purple colors are due to anthocyanin pigments produced in the outer layers of the kernel, the aleurone and pericarp. The genes that produce anthocyanin pigments are disabled (mutated) in yellow corn, allowing the yellow carotenoid pigments of the inner endosperm to show through. Both the anthocyanin and carotenoid pigment genes are mutated in white corn.
We can follow the inheritance of an anthocyanin pigment gene (called Red or R) simply by scoring the color of 100 kernels in an ear of corn, like the one in Figure 2. In this image, the phenotypic ratio of pigmented (dark red) to unpigmented (yellow) kernels is about 75:25, or 3:1.
Knowing just 4 facts about this genetic system, we can work backwards to figure out the genotype, or combination of alleles, present in each parent:
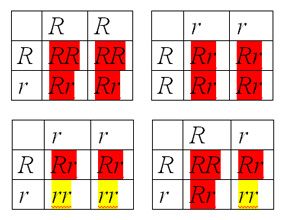 |
Figure 3 Punnett squares can predict the ratio of pigmented to unpigmented kernels. |
Using Punnett squares (Figure 3) we can predict what sorts of kernels (offspring) are produced by parents with different genotypes. We can try different combinations of parents with RR, Rr, or rr genotypes, as in Figure 3. Only crosses between 2 parents with the Rr genotype are expected to produce a 3:1 ratio of pigmented to unpigmented kernels.
A closer look reveals a more complicated story: some of the dark pigmented kernels are actually speckled with yellow (see Figure 4). This suggests that the pigment gene is not working properly in all of the cells in these kernels. What is going on? Something else seems to be modifying the activity of the R gene. A quick count reveals that about 1/3 of pigmented kernels are speckled. This is consistent with a modified R* gene that fails to express anthocyanin pigment in some cells, giving rise to yellow (recessive) sectors in heterozygous plants (R*r). Inactive regions are compensated by the active R allele in R*R kernels. In this case, the modification is believed to be in the promoter area that regulates the expression of the R gene. The modifcation * is not a change (mutation) in the DNA sequence itself. Rather, it is the addition of methyl groups (CH3), which block the binding of transcription factors needed to express the R gene consistently—thus turning it off in some cells.
 |
Figure 4 A closer look at pigmented kernels reveals yellow speckles. |
The pattern of DNA methylation is stably inherited through at least several generations. Because the methyl groups are “added to” or “on top of” the DNA sequence, this is called an epigenetic effect, and the methylated version of the gene is called an “epiallele.” “Imprinting” is the inheritance of an epiallele exclusively through the maternal or paternal line. In this case, the imprinted R* epiallele is inherited from the male pollen.
There is evidence for at least 80 imprinted genes in the human genome, some of which give rise to puzzling phenotypes. In one famous study, the grandsons of men who had survived starvation in the Swedish village of Norbatten lived on average 32 years longer than the grandsons of men who were well-fed during the same periods of famine. Starvation appears to have induced an epigenetic change that was inherited through generations to increase the fitness of the grandchildren. This interaction between genes and the environment would have been dismissed as Lamarckism only several decades ago.
In the next Carolina Tips® article in this series, we will take a close look at an epigenetic “switch” in the model plant Arabidopsis.