My Cart
Your Shopping Cart is currently empty. Use Quick Order or Search to quickly add items to your order!
 Anthony Eckdahl
Anthony Eckdahl
Junior, Central High School
St. Joseph, MO
August 2015
I have been interested in science since I can remember. Entering high school, I knew that I wanted to pursue my interest by taking biology, chemistry, and physics courses, and by doing a science fair project. After taking biology, I wanted to focus my project on some aspect of genetics and molecular biology. I also wanted my project to be one that let me collect and analyze original data. The pClone Red system allowed me to conduct authentic research on bacterial gene expression and to collect my own data.
Before I began my science project, my background in molecular genetics was minimal. I only had a basic understanding of it from freshman high school biology. However, I learned a lot of basic biology for my science fair project.
I learned that gene expression is the process by which the information of genes is used to direct the function of cells. Gene expression is regulated in all cells because not all genes are needed all the time or under all circumstances. For example, brain cells need to express certain genes that are not needed in muscle cells, and muscle cells need to express certain genes that are not needed in brain cells. Likewise, bacteria need to express different genes depending on the availability of food and other aspects of their surroundings.
I also learned that the regulation of gene expression can be used to manipulate cells for applications in pharmaceutical production, biofuels, production of useful materials, and more. I found all of this fascinating and was excited to learn more by doing a research project (Fig. 1).
Figure 1 Anthony Eckdahl uses the pClone Red system to conduct bacterial promoter research.
Gene expression involves the transcription of the DNA information of a gene into RNA and the translation of RNA information into the sequence of amino acids in a protein. RNA polymerase is an enzyme that transcribes DNA into RNA, and a promoter is a sequence of base pairs that is recognized by the RNA polymerase. In bacteria, transcription is initiated when the sigma factor and RNA polymerase recognize the -35 region of a promoter located upstream of a gene. The consensus sequence for the -35 region of E. coli promoters has been reported widely as TTGACA. RNA polymerase then interacts with the -10 region of the promoter and this results in separation of the 2 strands of DNA. RNA polymerase attaches itself to 1 of the 2 strands and begins to use that strand as a template to make RNA. RNA polymerase slides along the strand for the entire length of the gene and transcription ceases when RNA polymerase encounters a transcriptional terminator.
For my project, I decided to introduce mutations into the -35 region of a high efficiency promoter called Ptac and measure the effects of these mutations on gene expression. The pClone Red system allowed me to perform my promoter research without the need of expensive machinery or extensive training. According to the pClone Red procedure, I designed and ordered synthetic DNA with mutations in the -35 region that convert it from TTGACA to NNNNNN, where N represents any of the 4 DNA bases. I used Golden Gate Assembly (GGA) to make a library of mutant promoters in the pClone Red plasmid and introduced the library into E. coli. I spread the library of clones on agar plates, incubated them overnight, and took pictures on a UV box. I was able to produce a beautiful library of colonies, as seen in Fig. 2.
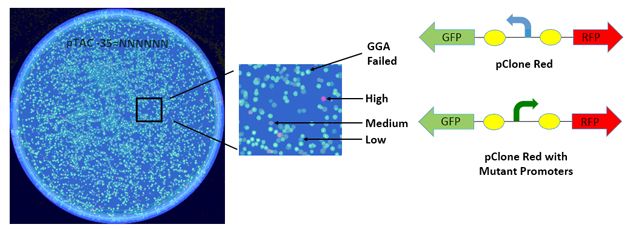
Figure 2 Colonies resulting from cloning Ptac promoter mutations can be distinguished in the pClone system from those resulting from failed Golden Gate Assembly.
As illustrated in Fig. 2, pClone Red allowed me to use a cloning method called Golden Gate Assembly to replace a left facing promoter that results in the production of green fluorescent protein with my mutated right facing promoters that promote various levels of red fluorescent protein. About half of my colonies were green, indicating that GGA had failed. The other half of the colonies had various shades of red fluorescence, indicating the effects of various mutations in the -35 region on promoter strength. I collected 81 colonies, purified the pClone Red plasmids from them, and had their DNA sequences determined. I also used a fluorimeter to measure the red fluorescence of cells from each of the 81 colonies.
The strengths of the promoters covered a wide range. Some were stronger than the original Ptac promoter, which surprised me. Some were similar to Ptac and others barely functioned. I determined my own consensus sequence for the -35 region and found that it was different from the sequence published in research papers and textbooks. It was exciting to get a result that was different from published results. I tested my consensus sequence by cloning 4 variations of it into pClone Red and showed that the consensus functioned with an efficiency similar to that of the published consensus. I concluded that the DNA sequence upstream and downstream of the -35 region has a bigger impact on promoter strength than was presented in textbooks or the scientific literature.
My research results provide insight into how particular mutations in the -35 region affect the overall efficiency of the Ptac bacterial promoter. The results of my study also provided information on a large collection of newly cloned promoter mutants and gene expression data for them. In synthetic biology research, the set of 81 promoter sequences of varying strengths can be a very valuable asset. These sequences could be used to tune promoter strength in devices with applications in biofuels, medicine, or the environment. In order to share the collection of 81 new promoters with the synthetic biology community, I contributed a page to the Registry of Standard Biological Parts, the largest collection of synthetic biology DNA parts in existence. I also entered information about my promoters into the Registry of Functional Promoters, a database that undergraduate and high school students can use to share the results of their pClone Red experiments.
I gave an oral presentation of my project at the regional 2015 Missouri Junior Academy and was invited to present again at the state level. I also presented my project in poster form at the Mid-America Regional Science and Engineering Fair, where I won the Grand Award and a trip to present at the 2015 International Science and Engineering Fair in Pittsburgh, PA.
I feel very fortunate to have had this success with my research project. None of it would have been possible without the support of my high school teacher, my host lab, and the pClone Red system.